Milk is rich in nutrients and contains fat, protein, minerals, vitamins, carbohydrates, and many other nutrients. It is a perfect environment for bacterial cultures, such as Salmonella, to grow due to its rich nutritional conditions. Unprocessed milk that has not been heat-treated or pasteurized can harbor a variety of bacteria that can cause Gastroenteritis and other intestinal diseases (Reference 1). Figure 1 shows the variety of bacterial cultures that could exist in milk.

Figure 1 : Différentes bactéries dans le lait
Another source of bacterial infection is poultry. The number of Salmonella cases in the United States exceeds 1 million per year, of which 200,000 originate from poultry alone (Reference 2). The bacterial infection can also occur in commercial fruit pulp such as pineapple. In addition, orange and mango juices can also be contaminated with bacteria (Reference 3).
The analytical methods of detection of Salmonella such as Polymerase Chain Reaction (PCR) require sample preparation, colony isolation and confirmation. A considerable amount of liquid and solid media needs to be consumed, and reagents are required. Not to mention, the test turnaround time is approximately 5 to 7 days.
A faster, non-destructive, non-invasive method of testing that does not require chemical reactions is required for Salmonella testing and identification. There are two candidates for this kind of fast testing. These two methods are hyperspectral imaging and near-infrared spectroscopy (NIRSImagerie microscopique (IHM) utilisant un filtre acousto-optique accordable (AOTF) et un microscope pour observer des cellules bactériennes individuelles issues de microcolonies sur des plaques d'agar, ainsi que l'analyse en composantes principales (ACP) pour détecter et identifier différents types de bactéries (référence 2). Une image de ce dispositif est montrée à la figure 2.

Figure 2 : Système d'imagerie microscopique AOTF pour détecter Salmonella (Référence 2)
The AOTF microscopic system classifies potential pathogens using only a few cells and analyzes the data using Chemometrics.
The second option for identifying Salmonella and possibly distinguishing it from other samples infected with the E. coli is Near-IR Spectroscopy (NIRS). The absorption in the near-IR region between 780-2500 nm consists of overtones and combination bands, which are broader and have lower resolution than the mid-IR region, where the fundamental absorptions occur. However, there is no need for sample preparation, and the measurements will take only a few seconds rather than a few days. Due to the complexity of spectra and the need to distinguish overlapping absorption bands, NIRS requires chemometric methods for data analysis. Several different bonds, such as C-H, N-H, O-H, and C=O, can be detected using NIRS. In one study, a near-IR spectrometer operating in the 900-1700 nm range was used to discriminate between Salmonella-infected and non-infected milk samples (Reference 1). After performing a PLS-DA (Partial Least Squares-Discriminant Analysis) examination of the second derivative of spectra, the following plot was obtained, as shown in Figure 3.
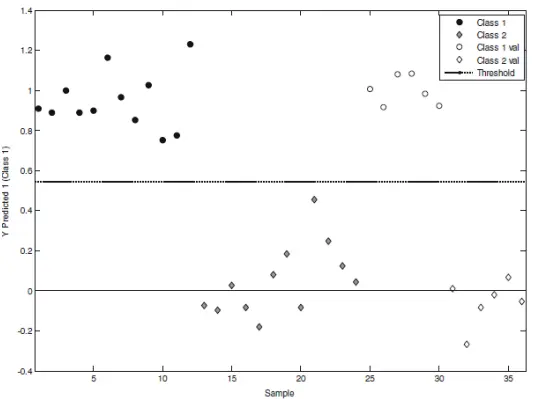
Figure 3 : La méthode PLS-DA distingue les échantillons contaminés et non contaminés dans le lait après
In the plot above, class 1 and class 2 refer to calibration samples (18 contaminated and 18 clean). Class 1 val and 2 val refer to validation samples (8 contaminated, 8 clean). It is clear that the two classes have been separated effectively using this method, and it demonstrates that NIRS enables real-time monitoring and can significantly reduce processing time.
In another study, the goal was to distinguish between fruit juice samples infected with Salmonella and those infected with E. coli (Reference 3). The Partial Least Squares (PLS) method failed to differentiate between the two types of bacteria, whereas PLS-DA clearly separated them. The measurement used an FT-NIR spectrometer over 1000-2500 nm to obtain the spectra. FT-NIR uses a Fourier transform technique and a near-infrared halogen Tungsten lamp to do the measurements. Figure 4 shows the comparison between the PCA method and the PLS-DA (Partial Least Squares-Discriminant Analysis) method. It seems that the former has failed and the latter has succeeded in distinguishing between Salmonella and E. coli.
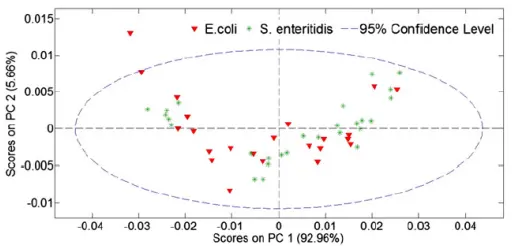
PLS-D
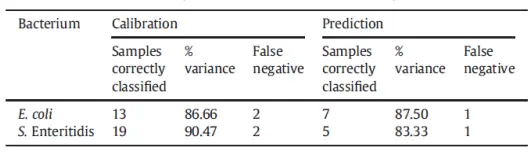
Figure 4 : Comparaison entre PCA et PLS-DA
So it becomes apparent that a proper chemometric technique is required for classification, and not every method can distinguish between samples of different natures.
Comparaison de différentes méthodes
In this blog, three different techniques were reviewed. Hyperspectral Microscopic Imaging (HMI), NIRS using a diffraction grating-based near-IR spectrometer, and finally NIRS using FT-NIR. All three methods used chemometrics to identify the bacteria. However, the hardware costs are high: both the HMI and FT-NIR are costly instruments priced in the tens of thousands of dollars. Diffraction grating-based NIR spectrometers can cost as little as $2000. Of course, the chemometric part of the work is entirely separate, and either needs to be done on a PC with statistical software such as MATLAB PLS Toolbox, or an app needs to be developed that uses cloud-based statistical tools, such as Python-based statistical libraries.
Allied Scientific Pro offers the Nirvascan spectrometer that comes in different models (Reflectance model for solids, Transmission model for liquids and fiber optic model which requires an external source). Although the spectrometer is a diffraction grating-based instrument, it uses Digital Light Processing (DLP) technology and a single-element InGaAs detector, which significantly reduces the price. A pic of each of the three models is shown in Figure 5.
Figure 5 : Différents modèles de spectromètre Nirvascan
Pour de plus amples renseignements, veuillez consulter le lien suivant.
https://alliedscientificpro. com/nirvascan
Références :
1- Discrimination rapide du lait contaminé par Salmonella sp. par spectroscopie proche infrarouge, J.M. Pereira et.al, Food Anal. Méthodes (2018).
2- Classification des sérotypes de Salmonella par imagerie au microscope hyperspectral, B. Park et al., Ann Clin Pathol 5(2):1108 (2017)
3- L'utilisation de la spectroscopie proche infrarouge et de techniques multivariées pour différencier Escherichia coli et Salmonella Enteritidis inoculés dans du jus de pulpe, A. Marques et.al, Journal of Microbiological methods 93 (2013) 90-94
